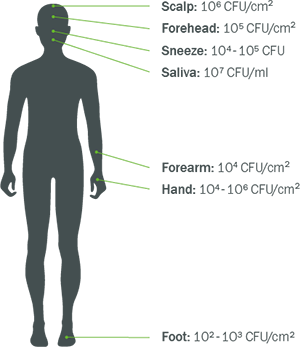
The MAIN SOURCE of M. fermentans found in tissues and samples, is from the HUMAN SCIENTISTS who BREATHED on the SAMPLES and introduced the contamination themselves
AND how some of these contaminants also exist in the PCR KITS
The major source of mycoplasma contamination is laboratory personnel.
M. fermentans exists on different parts of the human body including the mouth and the nose.
(Doctors/Scientists MAY be talking about this right now on X, in certain “circles”)
This means that if M. fermentans was detected in tissues or fluid samples, there is a high probability, it came from the scientist who was testing the samples—it has a far less likely chance that the presence of M. fermentans already existed in the sample itself. It was never there, until the scientist doing the testing, BREATHED on it.
The scientist could have coughed, sneezed or breathed on the samples they were testing, which might have been free of M. fermentans, but because it exists on all of us, it is easy to accidentally be transferred from the scientist to the sample, meaning the tests that show tissue or fluids contaminated with M. Fermenans, are most likely, FALSE POSITIVES.
(Meaning—the M. fermentans was never in the sample, until the scientist BREATHED on the sample).
The majority of species contaminating cell cultures are of human origin, including:
Mycoplasma orale, Mycoplasma fermentans, and Mycoplasma hominis.
Contamination occurs via aerosols generated by sneezing, coughing, or even talking, which can settle onto cell cultures or lab surfaces. Dirty lab coats and street clothes further contribute by shedding dust particles harboring mycoplasmas.
This can actually spread RAPIDLY across cell cultures.
”Once you have a mycoplasma-infected cell culture in your lab, it can spread among all other cultures via cross-contamination. Operator-induced contamination is a multifaceted problem. Mycoplasmas are spread by using laboratory equipment, media, or reagents that have been contaminated by previous use in processing mycoplasma-infected cells. Also incubators, water baths and other devices are often shared among multiple users and cell culture experiments, increasing the risk of cross-contamination between different cell lines.
It was shown, that in laboratories with contaminated cells, most or all cultures are positive containing the same mycoplasma species [4]. This is due to the ease of droplet generation during handling of cell cultures, the high concentration of mycoplasmas in infected cultures, and the prolonged survival of mycoplasma cells.”
Source: https://news.minerva-biolabs.com/informative-articles/2024/03/#:~:text=The%20major%20source%20of%20mycoplasma,other%20cultures%20via%20cross%2Dcontamination
That means that activities like pipetting, vortexing, and centrifugation generate aerosols capable of carrying mycoplasmas over considerable distances. Even in well-designed laminar flow hoods, improper technique can spread contamination. For instance, McGarrity’s study demonstrated that live mycoplasmas remained on hood surfaces 4–6 days post exposure, contaminating clean cultures subcultured weeks later.
Also:
Occurrence of Fungal DNA Contamination in PCR Reagents: Approaches to Control and Decontamination
”Problems with fungal contamination during PCR-based assays have been previously reported. Airborne conidia or spores and trace amounts of fungal nucleic acids in commercially available enzymes used for fungal cell lysis and in components of DNA extraction kits were identified as possible sources of contamination (7–10). Here we provide the first evidence for the presence of fungal DNA contaminants in lyophilized primers and TaqMan probes as well as in master mix solutions for real-time PCR analysis purchased at renowned biotechnology companies. Using a TaqMan-based panfungal PCR assay (11) established for sensitive screening of immunocompromised patients at risk for IFI, we detected traces of fungal DNA in several batches of the indicated reagents. To overcome the risk of false-positive PCR test results attributable to this issue, we have successfully applied a decontamination method based on the activity of a double-strand specific DNase (dsDNase).”
RESULTS
Identification of fungal DNA contamination in PCR reagents.
Despite the rigorous measures for the prevention of fungal contamination described above, false-positive results of panfungal PCR analysis were revealed by negative controls in a high proportion of samples tested within individual series (Table 1). The rate of false-positive test results was evaluated by performing nine independent series of negative-control reactions (n = 40 to 92 per series) in which DNA-free water replaced the template (no-template controls [NTC]), and the entire sample processing was performed using the stringent precautions outlined above. Within individual series of NTC specimens tested by panfungal PCR analysis, false-positive results were observed in up to 26.7% of samples analyzed (Table 1). The frequency of false-positive results remained constant as long as the same batches of master mix and primer and probe preparations were tested. Possible contamination with fungal DNA was determined by screening every new lot of individual reagents in PCR assays with components previously shown to be free of contaminants. Test results from two different batches of commercial master mix preparations are displayed (see Table 2). Batch 1 of the master mix was apparently free of fungal contamination as indicated by the consistently very low rate of false-positive signals in the presence of DNase-treated primers and probes. In contrast, batch 2 displayed an elevated level of false-positive signals in testing with treated oligonucleotides, thereby indicating the presence of traces of fungal DNA in the product. Nine independent test series, including a total of 532 NTC reactions, indicated that (i) both the oligonucleotides and master mixes provided the sources of contamination rather than other materials used and (ii) the levels of contamination with fungal DNA differed considerably between different batches of master mix and primer and probe solutions (Tables 1 and 2). Absence of any contamination with traces of fungal DNA, as determined by a highly sensitive panfungal PCR assay (11), was rarely observed in individual batches of these reagents obtained from different commercial providers. The panfungal PCR assay used includes two separate reactions primarily covering a broad spectrum of molds (reaction I) or yeasts and zygomycetes (reaction II) (11), and the majority of relevant contaminations were discovered by reaction II. To identify the fungal species involved, individual reagent components, including the primers, the TaqMan probe, and the master mix, were used as the template for amplification of the highly variable ITS2 region followed by Sanger sequencing analysis. The nucleotide sequences obtained showed high sequence similarities with the following fungal species derived from the GenBank database: Cryptococcus victoriae (99% identity within an overlap region of 195 bp), Malassezia restricta (99% identity within an overlap region of 407 bp), and Phlebia radiata (99% identity within an overlap region of 198 bp).”
That means that the fungus was present in the PCR test materials, used to test the samples themselves, and scientists might not realize the fungus came from the PCR test kits they used, and the fungus was not present in the sample.
The PCR kit itself, CONTAMINATED THE SAMPLE.
Others sources of contamination can be present in samples.
Coming soon: Review of the new (my) preprint (as soon as it goes live) raising potential concerns about two large studies and the methodologies used before analytics (meaning—review of two large studies, the methods the researchers used to work with the samples, the results of the tests, and why the methods may have potentially skewed the results—potentially and allegedly giving inaccurate information to the public, potentially their patients, why the lab tests for panels that are being charged for (people are paying $$$ for specific panels) might potentially and allegedly have concerns, and other consequences of these potentially inaccurate results.
I wrote the paper some time ago and was told not to publish it. I decided to upload it to the preprint server yesterday. Calling out concerns should not be silenced.
Layman’s explanations will be provided.
Sources:
Czurda, S., Smelik, S., Preuner-Stix, S., Nogueira, F., & Lion, T. (2016). Occurrence of Fungal DNA Contamination in PCR Reagents: Approaches to Control and Decontamination. Journal of clinical microbiology, 54(1), 148–152. https://doi.org/10.1128/JCM.02112-15
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., Turner, P., Parkhill, J., Loman, N. J., & Walker, A. W. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC biology, 12, 87. https://doi.org/10.1186/s12915-014-0087-z
Wally N, Schneider M, Thannesberger J, Kastner MT, Bakonyi T, Indik S, Rattei T, Bedarf J, Hildebrand F, Law J, Jovel J, Steininger C. Plasmid DNA contaminant in molecular reagents. Sci Rep. 2019 Feb 7;9(1):1652. doi: 10.1038/s41598-019-38733-1. PMID: 30733546; PMCID: PMC6367390.